We all are aware of the severeness of Cancer. It is estimated that nearly 18,000 adults die due to Brain Tumor and the survival rate tells us that if detected late then the person dies within the span of 5 years.
So, it is necessary that we devise a technique for early detection of the brain tumor and in today’s Modern World we have the power of AI to help us in the early diagnosis of these tumors.
In today’s world with the help of deep learning we can develop a Brain Tumor Detection app which can just by looking at your Brain CT scan would let you know the probability of you having Brain Tumor.
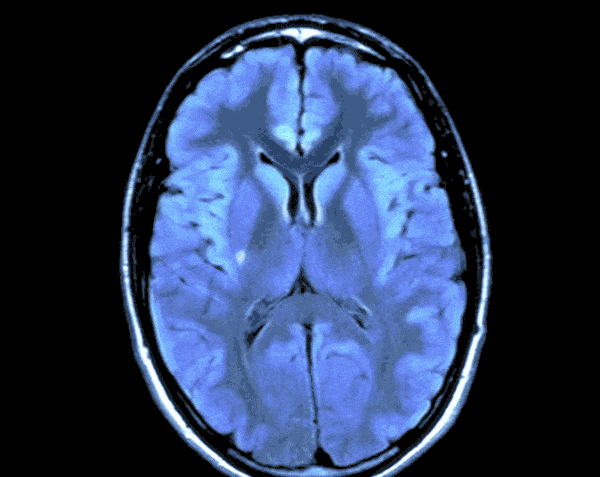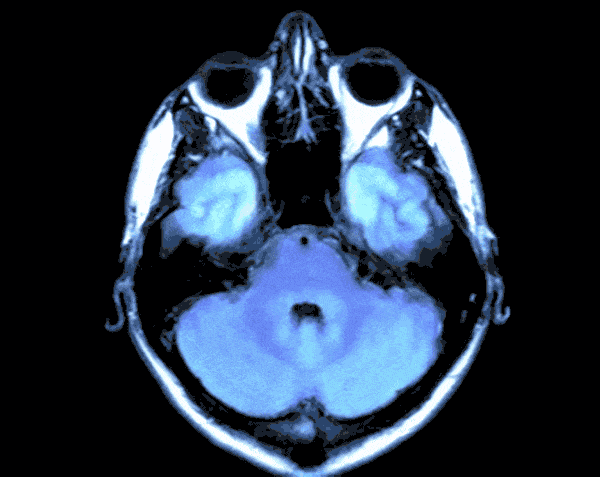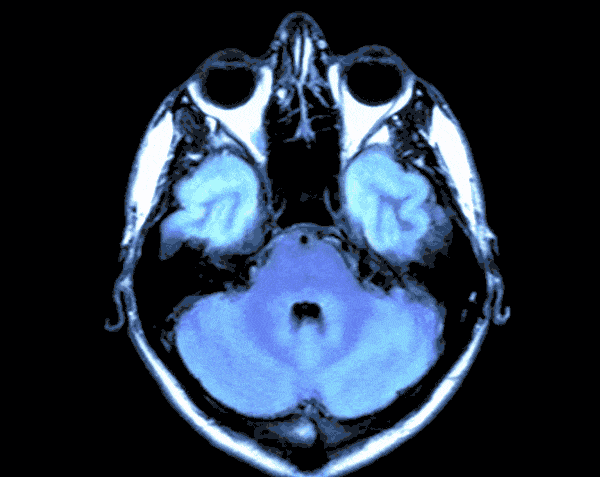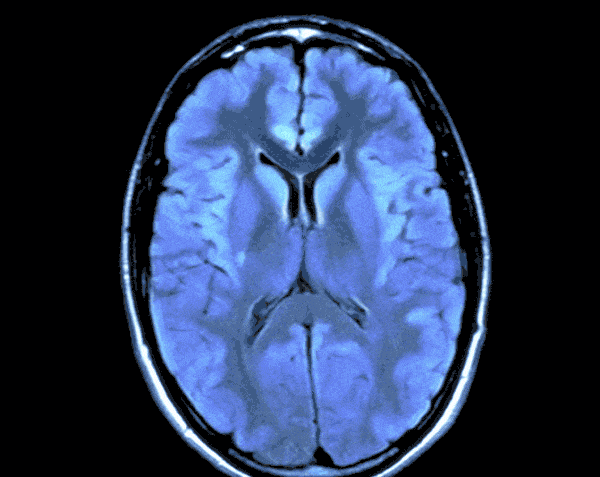
Table of Content
- Introduction to cAInvas
- Source of Data
- Data Augmentation
- Data Preprocessing
- Model Training
- Introduction to DeepC
- Compilation with DeepC
Introduction to cAInvas
cAInvas is an integrated development platform to create intelligent edge devices. Not only we can train our deep learning model using Tensorflow,Keras or Pytorch, we can also compile our model with its edge compiler called DeepC to deploy our working model on edge devices for production. The Brain Tumor Detection model which we are going to talk about, was developed on cAInvas.
cAInvas also offers various other deep learning notebooks in its gallery which one can use for reference or to gain insight about deep learning.It also has GPU support which makes it the best in its kind.
Source of Data
While working on cAInvas one of its key features is UseCases Gallary. When working on any of its UseCases you don’t have to look for data manually. As they have the feature to import the dataset to your workspace when you work on them. To load the data we just have to enter the following commands:
Running the above code in your notebook will load the labelled brain tumor data in your workspace.
Data Augmentation
It is generally suggested that while working on image data it is better to introduce some augmentations in the data like flipping the image, rotating the image by some angles, changing the brightness of the image, etc.
Since our deep learning model deals these images in the form of pixel values, so our model thinks these augmented data as new set of data and it improves the performance of the model. For image augmentations we can make a function and pass the image data through it and save them in our directory.
Data Preprocessing
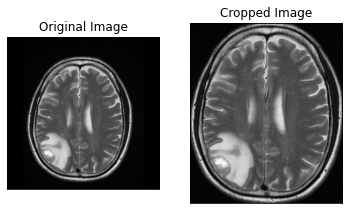
As the CT scans contain the images of the brain in the center surrounded by the blank area so in this step we would find the region that contains the image of the brain and crop the rest from the image. For this we would create a function.
By running the above function we will get the cropped image of the CT Scans and the results would look like this.
Data Loader and Model Training
Next step will be loading the data for model training. For this we can use OpenCV imread function and load the images as numpy array in one variable and their corresponding labels in another variable.Next, we will use scikit learn test-train split function for creating the train and test data.
Then we define the Model Architecture which is :
Model: "sequential" _________________________________________________________________ Layer (type) Output Shape Param # ================================================================= conv2d (Conv2D) (None, 218, 218, 32) 4736 _________________________________________________________________ bn0 (BatchNormalization) (None, 218, 218, 32) 128 _________________________________________________________________ max_pooling2d (MaxPooling2D) (None, 54, 54, 32) 0 _________________________________________________________________ max_pooling2d_1 (MaxPooling2 (None, 13, 13, 32) 0 _________________________________________________________________ flatten (Flatten) (None, 5408) 0 _________________________________________________________________ dense (Dense) (None, 1) 5409 ================================================================= Total params: 10,273 Trainable params: 10,209 Non-trainable params: 64 _________________________________________________________________
The loss function used was “binary_crossentropy” and optimizer used was “Adam”.For training the model we used Keras API with tensorflow at backend. The model showed good performance achieving a decent accuracy. Here are the training plots for the model:
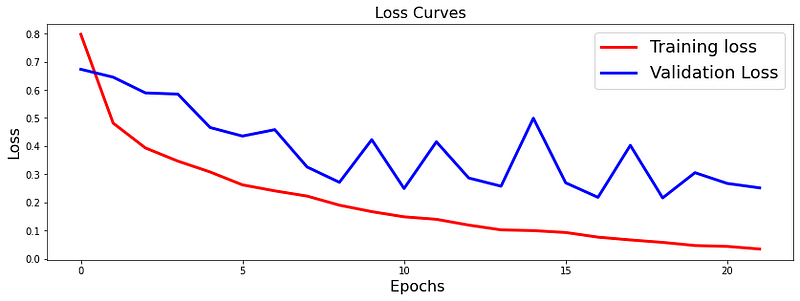
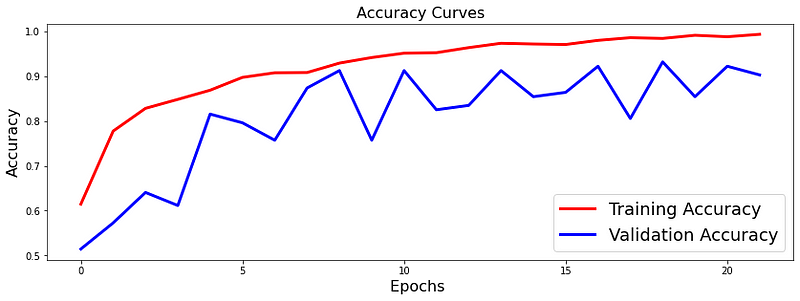
Also read: Skin Cancer Detection App
Introduction to DeepC
DeepC Compiler and inference framework is designed to enable and perform deep learning neural networks by focussing on features of small form-factor devices like micro-controllers, eFPGAs, cpus, and other embedded devices like raspberry-pi, odroid, Arduino, SparkFun Edge, risc-V, mobile phones, x86 and arm laptops among others.
DeepC also offers ahead of time compiler producing optimized executable based on LLVM compiler tool chain specialized for deep neural networks with ONNX as front end.
Compilation with DeepC
After training the model, it was saved in an H5 format using Keras as it easily stores the weights and model configuration in a single file.
After saving the file in H5 format we can easily compile our model using DeepC compiler which comes as a part of cAInvas platform so that it converts our saved model to a format which can be easily deployed to edge devices. And all this can be done very easily using a simple command.
And that’s all our Brain Tumor Detection model is ready for deployment.
Link for the cAInvas Notebook : https://cainvas.ai-tech.systems/use-cases/brain-tumor-detection-app/
Credit : Ashish Arya
You may also be interested in
- Learning more about Fingerprint pattern classification using Deep Learning
- Reading about Mushroom Classification Using Deep Learning
- Finding out about Deep Learning, the technology enabling machines to interpret complex data and mimic human cognition
Become a Contributor: Write for AITS Publication Today! We’ll be happy to publish your latest article on data science, artificial intelligence, machine learning, deep learning, and other technology topics
