
Introduction
Malaria is a life-threatening disease caused by parasites that are transmitted to people through the bites of infected female Anopheles mosquitoes. The World Health Organization states the following daunting facts about the disease on its website:
In 2019, there were an estimated 229 million cases of malaria worldwide. The estimated number of malaria deaths stood at 409 000 in 2019. Children aged under 5 years are the most vulnerable group affected by malaria; in 2019, they accounted for 67% (274 000) of all malaria deaths worldwide.
When it comes to battling diseases and development of healthcare facilities, Artificial Intelligence(AI) has been playing a major role and contributing heavily in the area. Today, AI offers a number of advantages over traditional medical practices and clinical decision making techniques.
These algorithms are becoming more and more accurate after observing huge amount of training data, providing unprecedented observations, care processes and patient outcomes.
The dataset
The malaria dataset provided by Lister Hill National Center for Biomedical Communications part of National Library of Medicine (NLM), contains 27,558 cell images with equal instances of parasitized and uninfected cells. The dataset consisted of colored images of the cells which were converted to grayscale and resized to a dimension of 50×50 to reduce the size of the images and overall dataset.
Each pixel has a single pixel-value associated with it, indicating the lightness or darkness of that pixel, with higher numbers meaning darker. This pixel-value is an integer between 0 and 255.
We have used binary classification in this project — a label of 0 means the cells are Parasitized and a label of 1 means the cells are Uninfected.
In this article, we will use Tensorflow — an opensource software library that provide tools and resources to create machine learning algorithms, and Keras — an interface for the Tensorflow library for developing deep learning models, to create a convolutional neural network and try to accurately predict whether the cells are parasitized or uninfected.
The entire code will be written on the Cainvas platform’s Notebook Server for better performance as well as scaling the model later to use it in EDGE devices.
Setting Up the Platform
You can create an account on the Cainvas website here.

After successful creation of an account, login to the platform and go to the Dashboard section to open the Notebook Server.
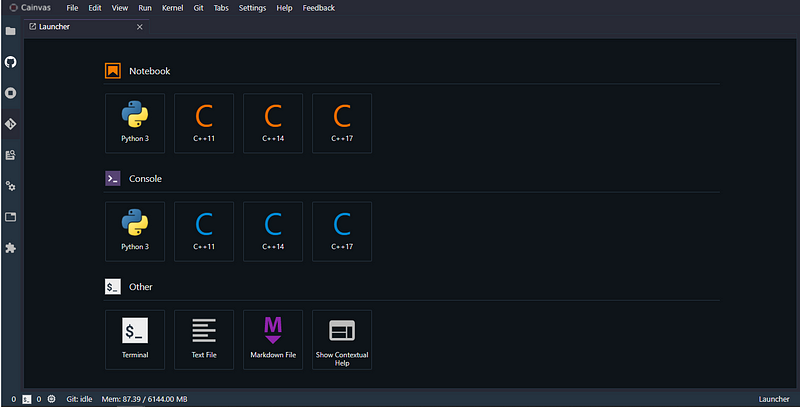
Importing the necessary libraries
We will use some commonly used libraries like Numpy and Matplotlib. We will use OpenCV2 and Matplotlib to access the images and display it in the notebook.
Other imports include Tensorflow and Keras to create the convolutional neural network and perform preprocessing of the data to perform training on it.
Loading the dataset
Cainvas platform allows us to upload datasets on the platform which facilitates ease of use. These datasets can be then easily loaded on to the notebook and used with enough flexibility to create the model without any hassle.
In order to upload your dataset, you can head to the Pailette section which allows uploading of files, images, videos and even sensor data.
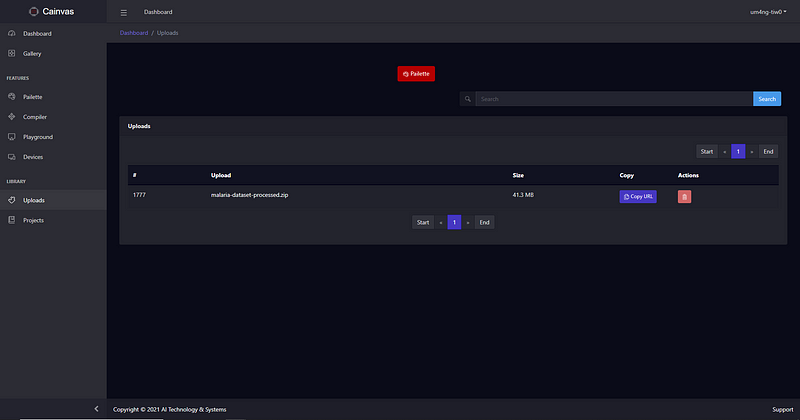
We will upload the dataset as a zip file in this article. The URL of the uploaded file can be obtained after the upload and used in the notebook to fetch it. To view the uploaded files just click on the Uploads sections. Click on the Copy URL button to copy the URL of the file.
We can use the URL with !wget command to load it in our notebook. We can then unzip the zip file in quiet mode using!unzip -qo filename.zip
We can access an image to check if the dataset has been loaded successfully.
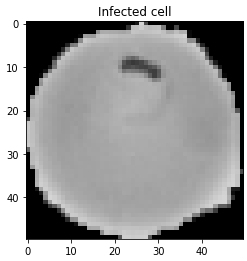
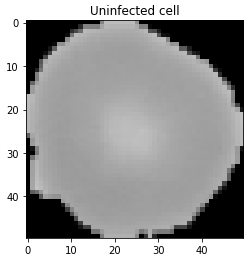
Let’s also access an uninfected cell image to see visual difference.
Preparing the data
The training data is present inside two folders — Parasitic & Uninfected. We will use the ImageDataGenerator offered by Keras to prepare the data and get appropriate labels pertaining to the folder structure. The generator also provides us the flexibility of creating train and validation split sets from the entire training dataset.
We can now check the labels created from the data.
array([0, 0, 0, ..., 1, 1, 1], dtype=int32)
As we can see, the labels are 0 and 1. The label 0 signifies the Parasitic cells and 1 signifies the Uninfected cells
Creating the Model
As mentioned, we will create a Convolutional Neural Network to predict the correct classes of cells from the images. We have used 3 Conv2D layers with MaxPool2D layers after each for the feature extraction from the images.
The activation function used is ReLU. The output layer has only one neuron, with Sigmoid activation function. The sigmoid activation function is a good choice in the output layer for binary classification applications.
We can now check the model summary
Model: "sequential" _________________________________________________________________ Layer (type) Output Shape Param # ================================================================= conv2d (Conv2D) (None, 48, 48, 16) 448 _________________________________________________________________ max_pooling2d (MaxPooling2D) (None, 24, 24, 16) 0 _________________________________________________________________ conv2d_1 (Conv2D) (None, 22, 22, 32) 4640 _________________________________________________________________ max_pooling2d_1 (MaxPooling2 (None, 11, 11, 32) 0 _________________________________________________________________ conv2d_2 (Conv2D) (None, 9, 9, 16) 4624 _________________________________________________________________ max_pooling2d_2 (MaxPooling2 (None, 4, 4, 16) 0 _________________________________________________________________ flatten (Flatten) (None, 256) 0 _________________________________________________________________ dense (Dense) (None, 32) 8224 _________________________________________________________________ dense_1 (Dense) (None, 1) 33 ================================================================= Total params: 17,969 Trainable params: 17,969 Non-trainable params: 0 _________________________________________________________________
We will use Early Stopping so that our model stops training if the monitored parameter does not change over time. This will make the training process more efficient
Compiling and Training the Model
We will compile the model with Adam as the optimizer and Binary Crossentropy as the loss function. We will train the model for 100 epochs with the callback. We will store the accuracy, loss, val_accuracy, and val_loss at each epoch in the history for plotting meaningful data later.
The last three epochs of the training phase:
Epoch 10/100 1034/1034 [==============================] - 5s 5ms/step - loss: 0.1216 - accuracy: 0.9565 - val_loss: 0.1591 - val_accuracy: 0.9430 Epoch 11/100 1034/1034 [==============================] - 5s 5ms/step - loss: 0.1157 - accuracy: 0.9569 - val_loss: 0.1706 - val_accuracy: 0.9388 Epoch 12/100 1034/1034 [==============================] - 5s 5ms/step - loss: 0.1086 - accuracy: 0.9606 - val_loss: 0.1677 - val_accuracy: 0.9419
Accuracy & Loss
We will plot the model performance at each epoch during the training phase.
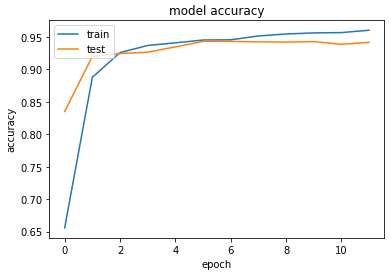
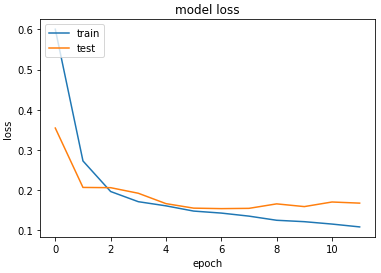
Testing the Model
We will now test the model by evaluating on the unseen test data which contains 4134 images.
259/259 [==============================] - 1s 4ms/step - loss: 0.1558 - accuracy: 0.9432 [0.15583141148090363, 0.9431543350219727]
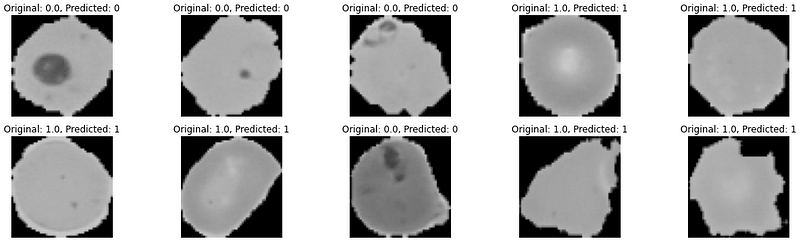
We will now predict the first 10 images in the test data.
Predicted Values: [0, 0, 0, 1, 1, 1, 1, 0, 1, 1] Actual Values: [0. 0. 0. 1. 1. 1. 1. 0. 1. 1.]
Visualizing the predictions for better insights.
Conclusion
In this article, we saw how to predict if a given cell is parasitized by malaria or is uninfected using a convolutional neural network created on the Cainvas Platform.
We observed the capabilities of artificial intelligence and a simple use case of how it can be employed to make healthcare systems smarter. We also saw the dataset upload capability of the Cainvas platform and how it makes creation of models simpler and faster.
The Cainvas Platform provides a one-stop solution to creating deep learning models which can also be compiled into EDGE device friendly models for using it in your IOT projects. The platform boasts of various other tools and resources to guide you for your next deep learning IOT project.
Link to the notebook on Cainvas: https://cainvas.ai-tech.systems/use-cases/malaria-parasite-detection-app/
Credit: Umang Tiwari
Also Read: Disaster Tweet Prediction on Cainvas
