Introduction
Tomato Disease Detection with CNN Architecture – The aim of this project is to identify various diseases on tomatoes based on their leaves. It is very important in agriculture to identify diseases immediately.
To detect the problem in real time, we develop Deep Learning model that can be installed on Embedded ML devices and it can used in greenhouses or by adding the model to some apps people can manually upload photo of tomato leaves and check how healthy it is.
Our model is able to find healthy tomatoes and 9 different diseases. The public dataset, which is available on Kaggle has been used to train and test the model.
Preprocessing
• Importing necessary libraries
• Downloading images from the server
• Reading and Preprocessing images
Our main folder contains 2 folders, ‘train’ for training images, ‘val’ for the test images. Each of these folders contains 10 folders, in turn. One is named ‘healthy’ that contains healthy tomato leaf images and other 9 folder folders contains tomato leaf images with diseases.
We have 1000 images in each category for training purpose and 1000 images (100 images for each) to test our model. We go through the main folder and list train and test folder directories, then we list names of 10 folders in ‘train’ and ‘val’ folders, remove ‘tomato’ name from there and get either ‘healthy’ label or corresponding disease name.
After that, we read images from each of these healthy/disease folders and we make sure that each file is in ‘jpg’ format. At the end, we separate images into r, g, b channels, normalize them and stack back together.
Output:
Let’s look at our categories to see disease names
Output:

Let’s visualize 2 images from each category with their corresponding labels
Output:

• Encoder and decoder functions
Since Machine Learning algorithms cannot identify texts, we need to feed our targets as numerical data. As we don’t have any priority through our categories, we use One Hot Encoding method to encode our labels.
For this purpose we create a dictionary where each label is key and has integer value representing column, then we create matrix with 10 column, each corresponding for one label and we go through the samples, take column value of that label from the dictionary and assign 1 that column.
The other function is decoder that does reverse: takes our predicted matrix, finds column with highest probability and return corresponding text label for that column.
•Encoding our train and test labels
Splitting our data into train and validation sets, data augmentation and training
• Data splitting
• Data augmentation
In images, leaves mainly lie either horizontally or vertically, therefore we need more data with flips for our model so that it can identify horizontal and vertical view of the same leaf. For this purpose, we use ImageDataGenerator from tensorflow and change horizontal_flip and vertical_flip arguments to True.
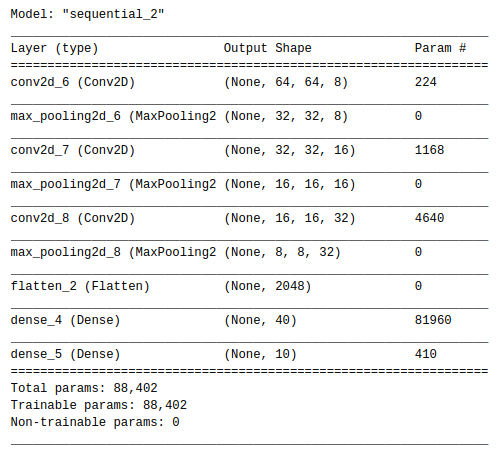
• Building CNN architecture
We use 3 blocks containing convolution layer, ReLU activation and Max Pooling layer for feature extraction and heading that flattens the results and use a Dense layer with 40 units, and use softmax activation for the final result
Output:
To train our model we use Adam optimizer with learning rate=0.0005, categorical crossentropy as loss function and accuracy as metric.
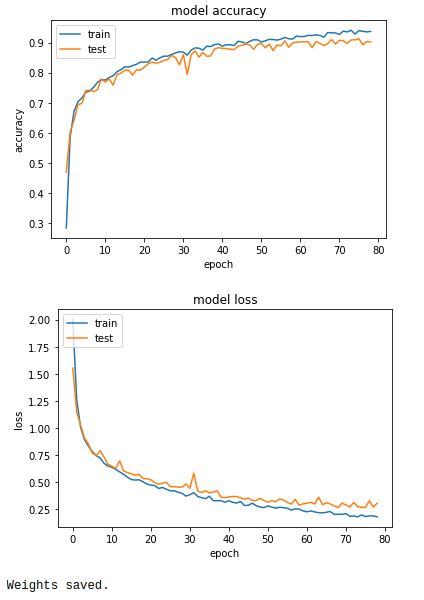
After training our model for 120 epochs, let’s look at how loss and accuracy changes over epochs and save our model. (During the train process callbacks have been used to monitor validation loss, if we don’t have an improvement for 10 epochs we use early stopping and create a check point)
Output:
Testing our model
To test our model, we use 100 images for each category. Let’s calculate our accuracy and loss on test images.
Output:

Visualizing some of the test results with their actual and predicted labels
Output:

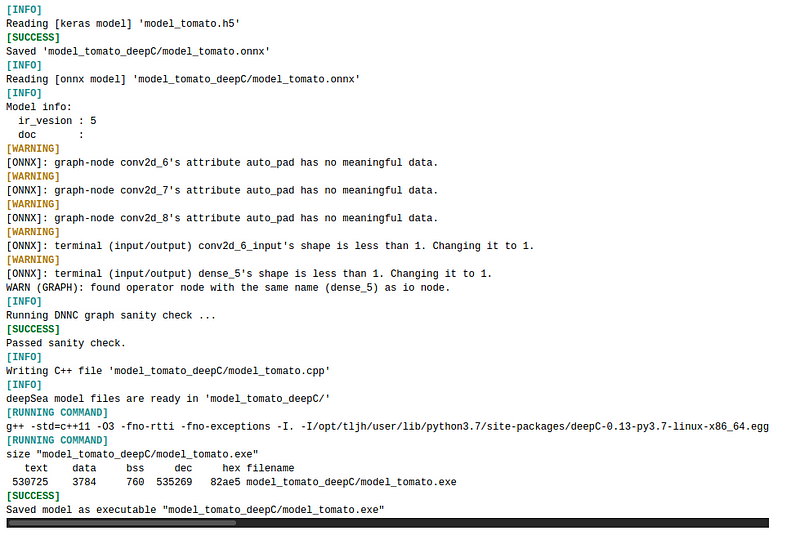
Transforming our model
We transform our model from ‘h5’ format to ‘exe’ format so that it will be ready to use on embedded devices
Output:
Conclusion
In conclusion, we build CNN architecture with 3 feature extraction blocks followed by a header to detect 9 diseases and healthy tomatoes based on their leaves. Due to data augmentation, our model is also able to identify flipped images and has high accuracy of over 90% on test data.
Notebook link — Here
Credit: Kanan Mahammadli
Also Read: Hand Sign Recognition with CNNs
