Classifying fetal health using CTG data in order to prevent child and maternal mortality.

United Nation’s Sustainable Development Goals reflect that reduction of child mortality is an indicator of human progress. This concept also includes maternal mortality. Most of the accounted losses have occurred in regions of low-resource and could have been prevented.
Cardiotocography (CTG)
Cardiotocography (CTG) is the means of measuring the fetal heart rate, movements, and uterine contractions, thus continuously monitoring the health of the mother and child.
The equipment used to perform the monitoring is called a cardiotocograph and work using ultrasound pulses. This is a simple and cost-effective solution for assessing fetal health, thus allowing professionals to take necessary action.
Implementation of the idea on cAInvas here!
The dataset
The dataset is a CSV file with 23 columns (22 attributes, 1 category column), and 2126 samples.
The features in the dataset are extracted from cardiotocograph exams and labeled by expert obstetricians into 3 classes — normal, suspect, and pathological.
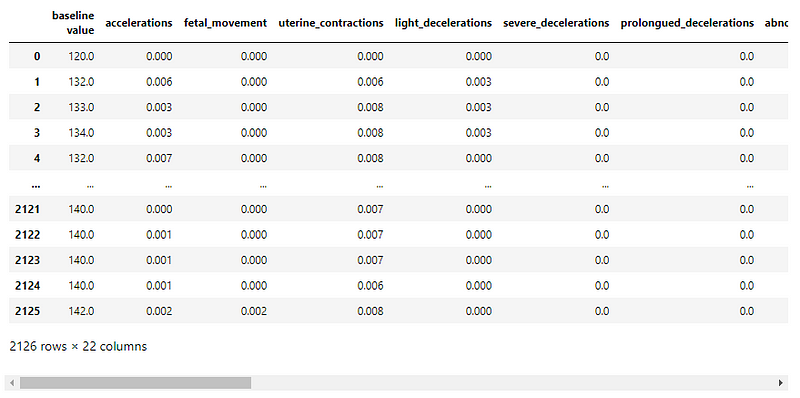
The columns in the dataset are
baseline value, accelerations, fetal_movement, uterine_contractions, light_decelerations, severe_decelerations, prolongued_decelerations, abnormal_short_term_variability, mean_value_of_short_term_variability, percentage_of_time_with_abnormal_long_term_variability, mean_value_of_long_term_variability, histogram_width, histogram_min, histogram_max, histogram_number_of_peaks, histogram_number_of_zeroes, histogram_mode, histogram_mean, histogram_median, histogram_variance, histogram_tendency, and fetal_health.
A peek into the spread of values across categories —
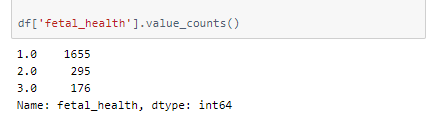
This is a heavily unbalanced dataset.
There are two ways to balance the dataset —
- Upsampling — Increasing the number of samples in a category by resampling them.
- Downsampling — Picking a subset of samples from the given set to use for training.
Here, we will be upsampling the categories to make their count equal to the class label with a higher count (here, 1655).
To start with, the data frame is divided into 3, one for each category.
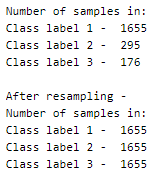
The three individual data frames are then concatenated into a single one that now has 4965 samples.
Preprocessing
Defining the input and output columns
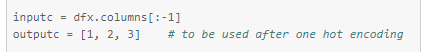
These column defining variables will be used to separate the train and test data frames into X and y (input and output).
One-hot encoding
The model’s outputs are one hot encoded as they use categorical cross-entropy.
Sample one hot encoding: Integer value 1 → [1, 0, 0], 2→[0, 1, 0], 3→ [0, 0, 1].
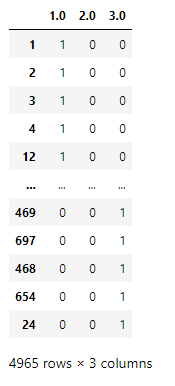
These one-hot encoded columns are added to the data frame.
Train-test split
Splitting into train and test sets after shuffling (as the category data frames are appended in order) using an 80–20 split ratio. This leaves 3972 samples in the training set and 993 samples in the test set. The data frames are then split into X and y using the column variables defined above.
Scaling the values
The attribute values are not of the same range. The MinMaxScaler function of the sklearn.preprocessing module is used to fit on the training set alone and use it to transform the values in both the train and test sets.
The model
The model is a simple one with 3 dense layers, two of which have ReLU activation and one has Softmax activation.
The model uses Adam optimizer and the categorical cross-entropy loss (as the category values are one-hot encoded). The accuracy of the predictions is tracked to evaluate the model’s performance.
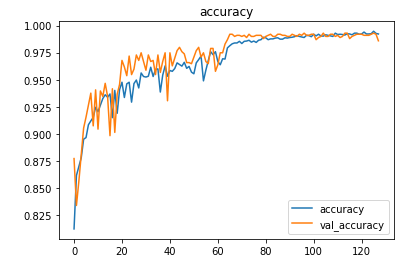
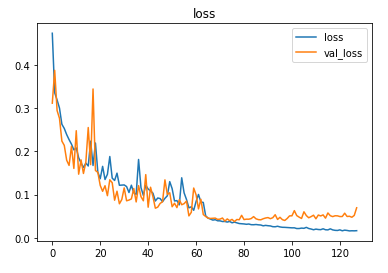
The model is trained for 64 epochs with a learning rate of 0.01 and another 64 epochs with 0.001 and achieved ~98.6% accuracy on the test set.
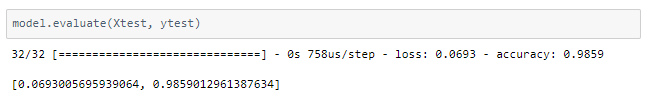
The metrics
Prediction
Let perform prediction on random test samples —

deepC
deepC library, compiler, and inference framework are designed to enable and perform deep learning neural networks by focussing on features of small form-factor devices like micro-controllers, eFPGAs, CPUs, and other embedded devices like raspberry-pi, odroid, Arduino, SparkFun Edge, RISC-V, mobile phones, x86 and arm laptops among others.
Compiling the model with deepC to get .exe file —
Head over to the cAInvas platform (link to notebook given earlier) and check out the predictions by the .exe file!
Credit: Ayisha D
You may also be interested in
- Learning more about Fingerprint pattern classification using Deep Learning
- Reading about Mushroom Classification Using Deep Learning
- Also Read: Next day rain prediction — on cAInvas
- Finding out about Deep Learning, the technology enabling machines to interpret complex data and mimic human cognition
Become a Contributor: Write for AITS Publication Today! We’ll be happy to publish your latest article on data science, artificial intelligence, machine learning, deep learning, and other technology topics
