According to a report of WHO, around 17.9 million people die each year due to Cardiovascular Diseases. Over the years it has been found that these deaths can be prevented if the diseases are diagnosed at an early stage and even the disease can be cured.

Table of Content
- AI Towards Healthcare
- Introduction to cAInvas
- Source of Data
- Data Analysis
- Feature Extraction
- Model Training
- Introduction to DeepC
- Compilation with DeepC
AI Towards Healthcare
Artificial Intelligence has been applied in various fields and one of them is AI for healthcare. We have seen AI practitioners coming up with solutions for various disease diagnosis such as Cancer Detection, Detection of Diabetic Retinopathy, and much more. The techniques used in these detections mostly involve Deep Learning.
So, by combining our knowledge of deep learning and with its integration Iot we can develop a smart digital-stethoscope which can help in diagnosing anomalies in heartbeat in real-time and can help in classifying Cardio-diseases.
Introduction to cAInvas
cAInvas is an integrated development platform to create intelligent edge devices. Not only we can train our deep learning model using Tensorflow, Keras, or Pytorch we can also compile our model with its edge compiler called DeepC to deploy our working model on edge devices for production. The Heartbeat Anomaly Detection model which we are going to talk about, was developed in cAInvas.
Source of Data
While working in cAInvas one of its key features is UseCases Gallary. When working on any of its UseCases you don’t have to look for data manually. As they have the feature to import your dataset to your workspace when you work on them. To load the data we just have to enter the following commands:
Data Analysis
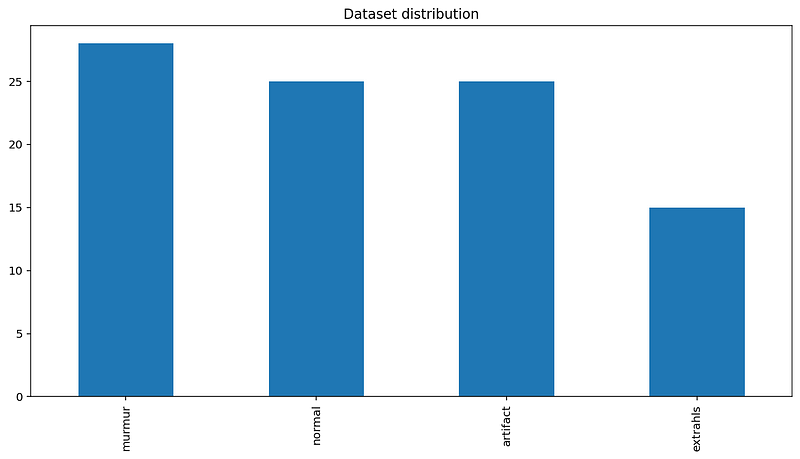
The dataset which we worked on consisted four kinds of heartbeat sounds namely:
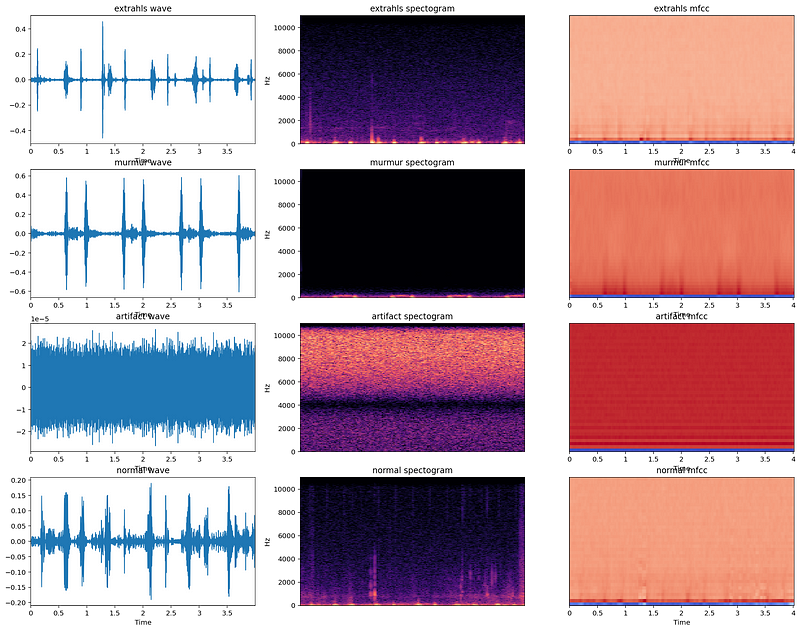
- Normal- In the Normal category there are normal, healthy heart sounds. A normal heart sound has a clear “lub dub, lub dub” pattern, with the time from “lub” to “dub” shorter than the time from “dub” to the next “lub”
- Murmur- Heart murmurs sound as though there is a “whooshing, roaring, rumbling, or turbulent fluid” noise in one of two temporal locations: (1) between “lub” and “dub”, or (2) between “dub” and “lub”. They can be a symptom of many heart disorders, some serious. There will still be a “lub” and a “dub”.
- Artifact- In the Artifact category there are a wide range of different sounds, including feedback squeals and echoes, speech, music and noise. There are usually no discernable heart sounds, and thus little or no temporal periodicity at frequencies below 195 Hz.
- Extrahls- Extrahl sounds may appear occasionally and can be identified because there is a heart sound that is out of rhythm involving extra or skipped heartbeats, e.g. a “lub-lub dub” or a “lub dub-dub”.It can be sign of a disease.
Let us see the Dataset Distribution:
Feature Extraction
As with all unstructured data formats, audio data has a couple of preprocessing steps which have to be followed before it is presented for analysis. Another way of representing audio data is by converting it into a different domain of data representation, namely the frequency domain.
There are a number of ways in which audio data can be represented such as using MFCs (Mel-Frequency cepstrums), Mel-spectrograms, and few more.
General Audio Features include:
- Time Domain features (eg. RMSE of waveform)
- Frequency domain features (eg. Amplitude of individual freuencies)
- Perceptual features (eg. MFCC)
- Windowing features (eg. Hamming distances of windows)
After extracting these features, it is then sent to the machine learning model for further analysis. For our model, we have used MFCC as our audio feature and that can be easily extracted from Audios using a python library called Librosa.To extract the features we just have to create a function and use librosa to extract MFCC features for us.
Model Training
After feature extraction, next step was to pass our extracted feature as training data for our Deep Learning model along with the target labels so that our model can learn to classify Heartbeat sounds and find anomalies in heartbeat sounds. The model architecture used was:
Model: "sequential" _________________________________________________________________ Layer (type) Output Shape Param # ================================================================= conv2d (Conv2D) (None, 39, 172, 16) 80 _________________________________________________________________ max_pooling2d (MaxPooling2D) (None, 19, 86, 16) 0 _________________________________________________________________ dropout (Dropout) (None, 19, 86, 16) 0 _________________________________________________________________ conv2d_1 (Conv2D) (None, 18, 85, 32) 2080 _________________________________________________________________ max_pooling2d_1 (MaxPooling2 (None, 9, 42, 32) 0 _________________________________________________________________ dropout_1 (Dropout) (None, 9, 42, 32) 0 _________________________________________________________________ conv2d_2 (Conv2D) (None, 8, 41, 64) 8256 _________________________________________________________________ max_pooling2d_2 (MaxPooling2 (None, 4, 20, 64) 0 _________________________________________________________________ dropout_2 (Dropout) (None, 4, 20, 64) 0 _________________________________________________________________ conv2d_3 (Conv2D) (None, 3, 19, 128) 32896 _________________________________________________________________ max_pooling2d_3 (MaxPooling2 (None, 1, 9, 128) 0 _________________________________________________________________ dropout_3 (Dropout) (None, 1, 9, 128) 0 _________________________________________________________________ global_average_pooling2d (Gl (None, 128) 0 _________________________________________________________________ dense (Dense) (None, 256) 33024 _________________________________________________________________ dense_1 (Dense) (None, 128) 32896 _________________________________________________________________ dense_2 (Dense) (None, 4) 516 ================================================================= Total params: 109,748 Trainable params: 109,748 Non-trainable params: 0 _________________________________________________________________
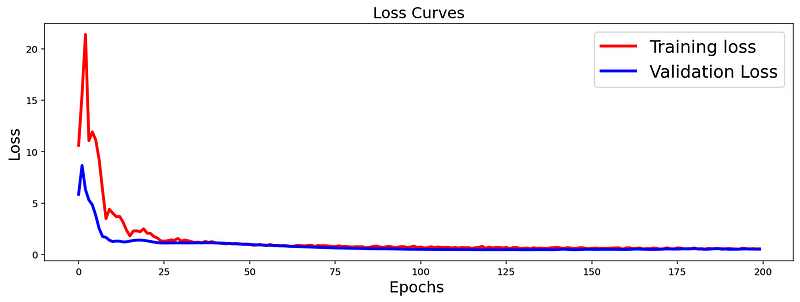
The loss function used was “categorical_crossentropy” and optimizer used was “Adam”.For training the model we used Keras API with tensorflow at backend. The model showed good performance achieving a decent accuracy. Here is the loss curve for the model:
Introduction to DeepC
DeepC Compiler and inference framework is designed to enable and perform deep learning neural networks by focussing on features of small form-factor devices like micro-controllers, eFPGAs, cpus, and other embedded devices like raspberry-pi, odroid, arduino, SparkFun Edge, risc-V, mobile phones, x86 and arm laptops among others.
DeepC also offers ahead of time compiler producing optimized executable based on LLVM compiler tool chain specialized for deep neural networks with ONNX as front end.
Compilation with DeepC
After training the model, it was saved in an H5 format using Keras as it easily stores the weights and model configuration in a single file.
After saving the file in H5 format we can easily compile our model using DeepC compiler which comes as a part of cAInvas platform so that it converts our saved model to a format which can be easily deployed to edge devices. And all this can be done very easily using a simple command.
And that’s all it is that easy to build a Heartbeat Anomaly Detection or any Deep Learning model with cAInvas.
Link for the cAInvas Notebook: https://cainvas.ai-tech.systems/use-cases/heartbeat-anomaly-detection-app-using-ecg/
Credit: Ashish Arya
You may also be interested in
- Learning more about Parkinson’s Disease Detection using Spiral Drawings and CNN
- Reading about Heart Disease Prediction using Neural Networks
- Also Read: ASL Recognition with TinyML devices
- Finding out about Artificial Intelligence, the brain-inspired algorithms revolutionizing how machines see and understand the world around us
Become a Contributor: Write for AITS Publication Today! We’ll be happy to publish your latest article on data science, artificial intelligence, machine learning, deep learning, and other technology topics
