Training a deep learning model to prescribe a drug based on the patient’s data.
A prescription drug is one that requires a medical prescription to be dispensed by law. On the other hand, an over-the-counter drug is one that can be dispensed without a prescription.
When it comes to the prescription of drugs, doctors look into various attributes of patient-related data before coming to a conclusion. This can have consequences varying from the efficiency of the medicine in the patient’s body to side effects caused and incorrect prescriptions may in some cases lead to irrevocable effects in patients (including death).
To start with, can we train a deep-learning model to prescribe medicines to patients based on their medical data? Read on to find out!
Implementation of the idea on cAInvas — here!
The dataset
The dataset is a CSV file with the features regarding a patient that affects drug prescriptions like age, sex, BP level, cholesterol, and sodium-potassium ratio and the corresponding drug prescribes in each case.
Preprocessing
Balancing the dataset
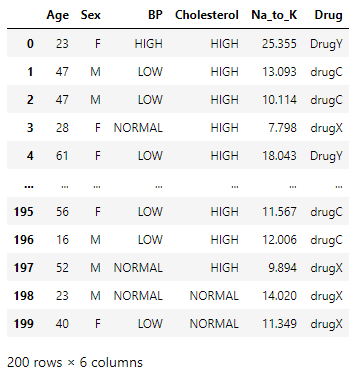
Looking into the classes and the spread of values among them in the dataset —
There are two ways to balance the dataset —
- upsampling — resample the values to make their count equal to the class label with the higher count (here, 91).
- downsampling — pick n samples from each class label where n = number of samples in class with least count (here, 16)
Here, we will be upsampling.
The replace parameter of .sample() is set to True to indicate that samples can be repeated in each class to achieve the given count. The df_balanced data frame has 455 samples, 91 of each class.
Categorical variables
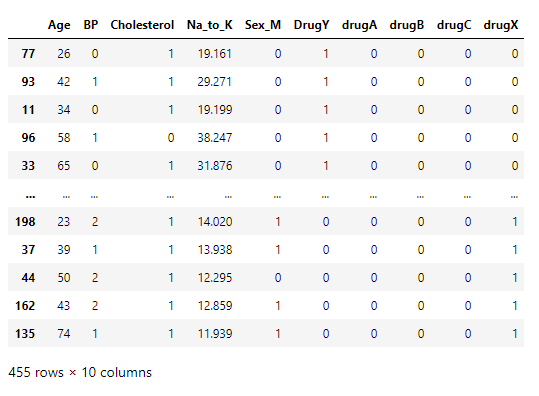
The ‘sex’ column does not define a range and thus is one-hot encoded while changing from a categorical attribute to a numerical attribute. This means if there are n unique values in the column, an array of length n is created for each where only the ith value is set to 1 with reference to an array that defines the indices of the column values in the array.
In many cases (mostly in the input columns), if there are n unique values, an array of length n-1 is created as the extra column can be redundant for identifying the column value from the encoded array. This is achieved by setting the drop_first parameter as True in the get_dummies() function as shown in the code cell below.
Since this column has only 2 unique values in the data frame, there will not be any difference between one-hot encoding and label encoding the column.
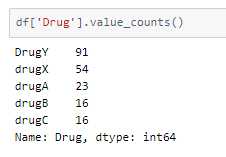
The values in the columns Cholesterol and BP represent range-kind values as seen by the values below.
These columns are label encoded instead of One-hot encoding, i.e, each value is replaced by a numeric value.
Since this is a classification problem, the output of the model which is now as an integer should be one-hot encoded.
Train-test split
Using an 80–10–10 ratio to split the data frame into train-validation-test sets. These are then divided into X and y (input and output) for further processing.
The training set has 364 samples while the validation set has 45 and the test set has 46 samples.
Scaling the values
A peek into the snapshot of the dfx data frame makes it evident that the columns have values in different ranges. The min-max scaler can be used to scale the values between the minimum and maximum values defined (min-0, max-1 by default).
The MinMaxScaler function of the sklearn.preprocessing module is used. Logically, the training set is the only data we are allowed to see or work with while training the model while the other two are used to evaluate its performance, the MinMaxScaler object is fit on the train data and the fitted model is used to transform the data in all the three datasets.
The model
The model is a simple one consisting only of Dense layers.
The model is compiled using the Cross-entropy loss function because the final layer of the model has the softmax activation function and the labels are one-hot encoded. The Adam optimizer is used and the accuracy of the model is tracked over epochs.
The EarlyStopping callback function monitors the validation loss and stops the training if it doesn’t decrease for 8 epochs continuously. The restore_best_weights parameter ensures that the model with the least validation loss is restored to the model variable.
The model is trained with a learning rate of 0.01 for 64 epochs but the model stops before that due to the callbacks.
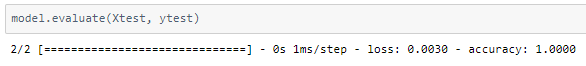
The model achieved 100% accuracy on the test set.
In problems such as these, it is important to keep the accuracy extremely high (100%) as chances cannot be taken with a patient’s medication.
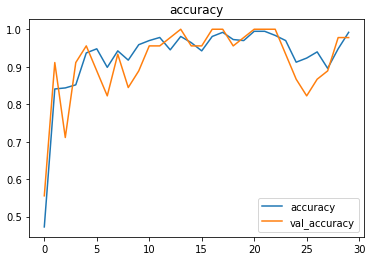
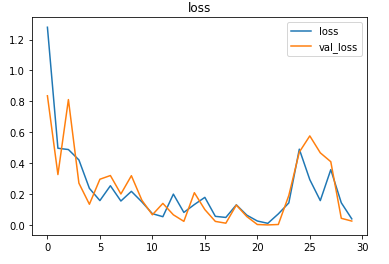
The metrics
Prediction
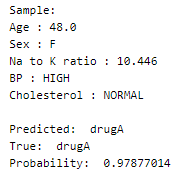
Let’s perform predictions on random test data samples —
Find the implementation of the print_sample() function in the notebook link above!
deepC
deepC library, compiler, and inference framework are designed to enable and perform deep learning neural networks by focussing on features of small form-factor devices like micro-controllers, eFPGAs, CPUs, and other embedded devices like raspberry-pi, odroid, Arduino, SparkFun Edge, RISC-V, mobile phones, x86 and arm laptops among others.
Compiling the model using deepC —
Head over to the cAInvas platform (link to notebook given earlier) and check out the predictions by the .exe file!
Credits: Ayisha D
Also Read: Sheep breed classification — on cAInvas
