Context
This database contains 76 attributes, but all published experiments refer to using a subset of 14 of them. In particular, the Cleveland database is the only one that has been used by ML researchers to
this date. The “goal” field refers to the presence of heart disease in the patient. It is integer-valued from 0 (no presence) to 4.
Content
Attribute Information:
- age
- sex
- chest pain type (4 values)
- resting blood pressure
- serum cholesterol in mg/dl
- fasting blood sugar > 120 mg/dl
- resting electrocardiographic results (values 0,1,2)
- maximum heart rate achieved
- exercise-induced angina
- old peak = ST depression induced by exercise relative to rest
- the slope of the peak exercise ST segment
- number of major vessels (0–3) colored by fluoroscopy
- thal: 3 = normal; 6 = fixed defect; 7 = reversible defect
Importing Libraries
Read Data
loading the data into the notebook
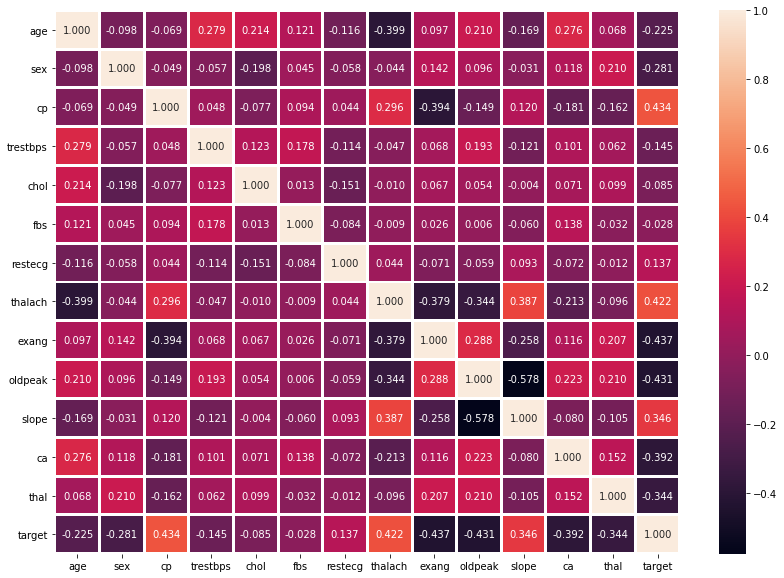
Data Analysis
Visualization of the data through graphs and heat maps
Data Preprocessing
Making Dummy columns
Index([\'age\', \'sex\', \'trestbps\', \'chol\', \'fbs\', \'restecg\', \'thalach\', \'exang\',
\'oldpeak\', \'ca\', \'target\', \'cp_0\', \'cp_1\', \'cp_2\', \'cp_3\', \'thal_0\',
\'thal_1\', \'thal_2\', \'thal_3\', \'slope_0\', \'slope_1\', \'slope_2\'],
dtype=\'object\')
age int64 sex int64 trestbps int64 chol int64 fbs int64 restecg int64 thalach int64 exang int64 oldpeak float64 ca int64 target int64 cp_0 uint8 cp_1 uint8 cp_2 uint8 cp_3 uint8 thal_0 uint8 thal_1 uint8 thal_2 uint8 thal_3 uint8 slope_0 uint8 slope_1 uint8 slope_2 uint8 dtype: object
Dropping Waste Column
Train-Test Split
Splitting the data set in 80–20 split
Model Architecture
Architecture using Neural Networks
Model: "sequential" _________________________________________________________________ Layer (type) Output Shape Param # ================================================================= dense (Dense) (None, 12) 264 _________________________________________________________________ dropout (Dropout) (None, 12) 0 _________________________________________________________________ dense_1 (Dense) (None, 2) 26 ================================================================= Total params: 290 Trainable params: 290 Non-trainable params: 0 _________________________________________________________________
Training the Model
Trained model using Adam Optimiser
Epoch 1/50 22/22 [==============================] - 0s 7ms/step - loss: 0.6330 - accuracy: 0.7311 - val_loss: 0.6195 - val_accuracy: 0.7143 Epoch 2/50 22/22 [==============================] - 0s 2ms/step - loss: 0.5644 - accuracy: 0.8349 - val_loss: 0.5751 - val_accuracy: 0.7253 Epoch 3/50 22/22 [==============================] - 0s 2ms/step - loss: 0.5021 - accuracy: 0.8679 - val_loss: 0.5326 - val_accuracy: 0.7363 Epoch 4/50 22/22 [==============================] - 0s 2ms/step - loss: 0.4519 - accuracy: 0.8774 - val_loss: 0.5008 - val_accuracy: 0.7802 Epoch 5/50 22/22 [==============================] - 0s 2ms/step - loss: 0.4095 - accuracy: 0.8679 - val_loss: 0.4799 - val_accuracy: 0.7692 Epoch 6/50 22/22 [==============================] - 0s 2ms/step - loss: 0.3760 - accuracy: 0.8632 - val_loss: 0.4683 - val_accuracy: 0.7692 Epoch 7/50 22/22 [==============================] - 0s 2ms/step - loss: 0.3642 - accuracy: 0.8726 - val_loss: 0.4642 - val_accuracy: 0.7692 Epoch 8/50 22/22 [==============================] - 0s 2ms/step - loss: 0.3463 - accuracy: 0.8585 - val_loss: 0.4645 - val_accuracy: 0.7582 Epoch 9/50 22/22 [==============================] - 0s 2ms/step - loss: 0.3448 - accuracy: 0.8821 - val_loss: 0.4648 - val_accuracy: 0.7692 Epoch 10/50 22/22 [==============================] - 0s 2ms/step - loss: 0.3219 - accuracy: 0.8821 - val_loss: 0.4651 - val_accuracy: 0.7802 Epoch 11/50 22/22 [==============================] - 0s 2ms/step - loss: 0.3218 - accuracy: 0.9009 - val_loss: 0.4669 - val_accuracy: 0.7802 Epoch 12/50 22/22 [==============================] - 0s 2ms/step - loss: 0.3298 - accuracy: 0.8868 - val_loss: 0.4714 - val_accuracy: 0.7692 Epoch 13/50 22/22 [==============================] - 0s 2ms/step - loss: 0.3189 - accuracy: 0.8868 - val_loss: 0.4727 - val_accuracy: 0.7802 Epoch 14/50 22/22 [==============================] - 0s 2ms/step - loss: 0.3097 - accuracy: 0.8821 - val_loss: 0.4759 - val_accuracy: 0.7582 Epoch 15/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2999 - accuracy: 0.8962 - val_loss: 0.4785 - val_accuracy: 0.7802 Epoch 16/50 22/22 [==============================] - 0s 2ms/step - loss: 0.3017 - accuracy: 0.8962 - val_loss: 0.4840 - val_accuracy: 0.7692 Epoch 17/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2890 - accuracy: 0.8915 - val_loss: 0.4835 - val_accuracy: 0.7802 Epoch 18/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2858 - accuracy: 0.8962 - val_loss: 0.4903 - val_accuracy: 0.7582 Epoch 19/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2978 - accuracy: 0.8962 - val_loss: 0.4915 - val_accuracy: 0.7692 Epoch 20/50 22/22 [==============================] - 0s 2ms/step - loss: 0.3000 - accuracy: 0.8868 - val_loss: 0.4947 - val_accuracy: 0.7802 Epoch 21/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2750 - accuracy: 0.8962 - val_loss: 0.4994 - val_accuracy: 0.7692 Epoch 22/50 22/22 [==============================] - 0s 2ms/step - loss: 0.3057 - accuracy: 0.8962 - val_loss: 0.5014 - val_accuracy: 0.7802 Epoch 23/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2841 - accuracy: 0.8962 - val_loss: 0.5023 - val_accuracy: 0.7692 Epoch 24/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2750 - accuracy: 0.8962 - val_loss: 0.5018 - val_accuracy: 0.7692 Epoch 25/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2843 - accuracy: 0.9151 - val_loss: 0.5044 - val_accuracy: 0.7692 Epoch 26/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2800 - accuracy: 0.8868 - val_loss: 0.5091 - val_accuracy: 0.7692 Epoch 27/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2852 - accuracy: 0.9009 - val_loss: 0.5023 - val_accuracy: 0.7802 Epoch 28/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2617 - accuracy: 0.9009 - val_loss: 0.5051 - val_accuracy: 0.7692 Epoch 29/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2692 - accuracy: 0.8962 - val_loss: 0.5066 - val_accuracy: 0.7802 Epoch 30/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2681 - accuracy: 0.9009 - val_loss: 0.5142 - val_accuracy: 0.7692 Epoch 31/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2709 - accuracy: 0.9009 - val_loss: 0.5110 - val_accuracy: 0.7802 Epoch 32/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2801 - accuracy: 0.8868 - val_loss: 0.5114 - val_accuracy: 0.7912 Epoch 33/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2667 - accuracy: 0.8868 - val_loss: 0.5131 - val_accuracy: 0.7912 Epoch 34/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2741 - accuracy: 0.8868 - val_loss: 0.5107 - val_accuracy: 0.7802 Epoch 35/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2660 - accuracy: 0.9151 - val_loss: 0.5130 - val_accuracy: 0.7802 Epoch 36/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2781 - accuracy: 0.9009 - val_loss: 0.5132 - val_accuracy: 0.7802 Epoch 37/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2711 - accuracy: 0.9151 - val_loss: 0.5154 - val_accuracy: 0.7692 Epoch 38/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2584 - accuracy: 0.9104 - val_loss: 0.5148 - val_accuracy: 0.7802 Epoch 39/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2726 - accuracy: 0.8962 - val_loss: 0.5170 - val_accuracy: 0.7802 Epoch 40/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2684 - accuracy: 0.8962 - val_loss: 0.5225 - val_accuracy: 0.7802 Epoch 41/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2590 - accuracy: 0.9057 - val_loss: 0.5195 - val_accuracy: 0.7802 Epoch 42/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2608 - accuracy: 0.8962 - val_loss: 0.5241 - val_accuracy: 0.7802 Epoch 43/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2537 - accuracy: 0.9151 - val_loss: 0.5263 - val_accuracy: 0.7802 Epoch 44/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2485 - accuracy: 0.8962 - val_loss: 0.5251 - val_accuracy: 0.7802 Epoch 45/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2368 - accuracy: 0.9151 - val_loss: 0.5270 - val_accuracy: 0.7802 Epoch 46/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2573 - accuracy: 0.8962 - val_loss: 0.5306 - val_accuracy: 0.7802 Epoch 47/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2395 - accuracy: 0.9057 - val_loss: 0.5328 - val_accuracy: 0.7802 Epoch 48/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2634 - accuracy: 0.9104 - val_loss: 0.5322 - val_accuracy: 0.7802 Epoch 49/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2447 - accuracy: 0.9151 - val_loss: 0.5346 - val_accuracy: 0.7802 Epoch 50/50 22/22 [==============================] - 0s 2ms/step - loss: 0.2518 - accuracy: 0.9057 - val_loss: 0.5403 - val_accuracy: 0.7802
Accuracy and Loss Models
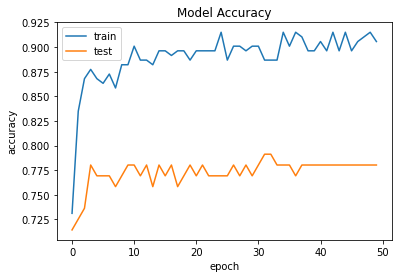
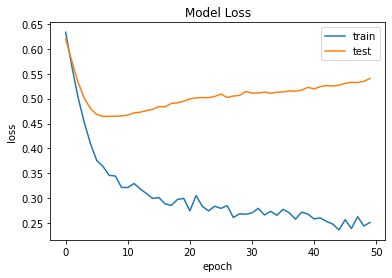
Result and Conclusion
precision recall f1-score support
0 0.78 0.71 0.74 41
1 0.78 0.84 0.81 50
micro avg 0.78 0.78 0.78 91 macro avg 0.78 0.77 0.78 91 weighted avg 0.78 0.78 0.78 91 samples avg 0.78 0.78 0.78 91
Saving the Model
array([1, 1, 0, 0, 1, 0, 1, 1, 1, 1, 0, 1, 1, 0, 1, 0, 0, 0, 1, 0, 1, 1,
1, 1, 1, 1, 0, 0, 1, 0, 0, 1, 1, 1, 0, 1, 1, 0, 1, 1, 1, 1, 0, 1,
1, 0, 0, 0, 0, 1, 1, 0, 1, 1, 1, 1, 0, 0, 1, 1, 0, 0, 1, 1, 1, 0,
1, 0, 0, 1, 1, 1, 1, 1, 1, 0, 1, 1, 0, 0, 1, 0, 1, 1, 1, 0, 1, 1,
1, 0, 0])
DeepCC
[INFO] Reading [keras model] \'heart_disease_ucl.h5\' [SUCCESS] Saved \'heart_disease_ucl_deepC/heart_disease_ucl.onnx\' [INFO] Reading [onnx model] \'heart_disease_ucl_deepC/heart_disease_ucl.onnx\' [INFO] Model info: ir_vesion : 4 doc : [WARNING] [ONNX]: terminal (input/output) dense_input\'s shape is less than 1. Changing it to 1. [WARNING] [ONNX]: terminal (input/output) dense_1\'s shape is less than 1. Changing it to 1. WARN (GRAPH): found operator node with the same name (dense_1) as io node. [INFO] Running DNNC graph sanity check ... [SUCCESS] Passed sanity check. [INFO] Writing C++ file \'heart_disease_ucl_deepC/heart_disease_ucl.cpp\' [INFO] deepSea model files are ready in \'heart_disease_ucl_deepC/\' [RUNNING COMMAND] g++ -std=c++11 -O3 -fno-rtti -fno-exceptions -I. -I/opt/tljh/user/lib/python3.7/site-packages/deepC-0.13-py3.7-linux-x86_64.egg/deepC/include -isystem /opt/tljh/user/lib/python3.7/site-packages/deepC-0.13-py3.7-linux-x86_64.egg/deepC/packages/eigen-eigen-323c052e1731 "heart_disease_ucl_deepC/heart_disease_ucl.cpp" -D_AITS_MAIN -o "heart_disease_ucl_deepC/heart_disease_ucl.exe" [RUNNING COMMAND] size "heart_disease_ucl_deepC/heart_disease_ucl.exe" text data bss dec hex filename 122165 2984 760 125909 1ebd5 heart_disease_ucl_deepC/heart_disease_ucl.exe [SUCCESS] Saved model as executable "heart_disease_ucl_deepC/heart_disease_ucl.exe"
This is how we make a model for the prediction of a heart disease dataset using Neural Networks. There are a million ways to make a model because of recent and upcoming developments in the field of Deep Learning.
Notebook Link: Here
Credits: Siddharth Ganjoo
Also Read: Tomato Disease Detection with CNN Architecturereplica patek philippe vintage
